Unbounding the Future:
the Nanotechnology Revolution
Chapter 4
Paths, Pioneers, and Progress
A basic question about nanotechnology
is, "When will it be achieved?" The answer is simple:
No one knows. How molecular
machines will behave is a matter for calculation, but how
long it will take us to develop them is a separate issue.
Technology timetables can't be calculated from the laws of
nature, they can only be guessed at. In this chapter, we examine
different paths to nanotechnology, hear what some of the pioneers
have to say, and describe the progress already made. This will
not answer our basic question, but it will educate our guesses.
Molecular
nanotechnology could be developed in any of several basically
different ways. Each of these basic alternatives itself includes
further alternatives. Researchers will be asking, "How can
we make the fastest progress?" To understand the answers
they may come to, we need to ask the same question here, adopting
(for the moment) a gung-ho, let's-go, how-do-we-get-the-job-done?
attitude. We give some of the researchers' answers in their own
words.
Will It Ever Be Achieved?
Like "When will it be achieved?", this is a basic
question with an answer beyond calculation. Here, though, the
answer seems fairly clear. Throughout history, people have worked
to achieve better control of matter, to convince atoms to do what we want them to
do. This has gone on since before people learned that atoms
exist, and has accelerated ever since. Although different
industries use different materials and different tools and
methods, the basic aim is always the same. They seek to make
better things, and make them more consistently, and that means
better control of the structure of matter. From this perspective,
nanotechnology is just the next, natural step in a progression
that has been under way for millennia.
Consider the compact discs now replacing older stereo records:
both the old and the new technologies stamp patterns into
plastic, but for CDs, the bumps on the stamping surface are only
about 130 by 600 nanometers in size, versus 100,000 nanometers or
so for the width of the groove on an old-style record. Or look at
a personal computer. John Foster, a physicist at IBM's Almaden
Research Center, points to a hard disk and says that "inside
that box are a bunch of whirring disks, and every one of those
disks has got a metal layer where the information is stored. The
last thing on top of the metal layer is a monolayer that's the
lubricant between the disk and the head that flies over it. The
monolayer is not fifteen angstroms [15 angstroms = 1.5
nanometers] and it's not three, because fifteen won't work and
neither will three. So it has to be ten plus or minus a few
angstroms. This is definitely working in the nanometer regime.
We're at that level: We ship it every day and make money on it
every day."
The transistors on computer chips are heading down in size on
an exponential curve. Foster's colleague at IBM, Patrick Arnett,
expects the trend to continue: "If you stay on that curve,
then you end up at the atomic scale at 2020 or so. That's the
nature of technology now. You expect to follow that curve as far
as you can go." The trend is clear, and at least some of the
results can be foreseen, but the precise path and timetable for
the development of nanotechnology is unpredictable. This
unpredictability goes to the heart of important questions:
"How will this technology be developed? Who will do it?
Where? When? In ten years? Fifty? A hundred? Will this happen in my
lifetime?" The answers will depend on what people do with
their time and resources, which in turn will depend on what goals
they think are most promising. Human attitudes, understanding,
and goals will make all the difference.
What Decisions Most Affect the Rate of
Advance?
Decisions about research directions are central. Researchers
are already pouring effort into chemical synthesis, molecular
engineering, and related fields. The same amount of effort could
produce more impressive results in molecular nanotechnology if a
fraction of it were differently directed. The research
funders—corporate executives, and decision makers in science
funding agencies like the National Science Foundation in the
United States and Japan's Ministry of International Trade and
Industry—all have a large influence on research directions,
but so do the researchers working in the labs. They submit
proposals to potential funders (and often spend time on
personally chosen projects, regardless of funding), so their
opinions also shape what happens. Where public money is involved,
politicians' impressions of public opinion can have a huge
influence, and public opinion depends on what all of us think and
say..
Still, researchers play a central role. They tend to work on
what they think is interesting, which depends on what they think
is possible, which depends on the tools they have or—among
the most creative researchers—on the tools they can see how
to make. Our tools shape how we think: as the saying goes, when
all you have is a hammer, everything looks like a nail. New tools
encourage new thoughts and enable new achievements, and decisions
about tool development will pace advances in nanotechnology. To
understand the challenges ahead, we need to take a look at ideas
about the tools that will be needed.
Why Are Tools So Important?
Throughout history, limited tools have limited achievement.
Leonardo da Vinci's sixteenth century chain drives and ball
bearings were theoretically workable, yet never worked in their
inventor's lifetime. Charles Babbage's nineteenth century
mechanical computer suffered the same fate. The problem? Both
inventors needed precisely machined parts that (though readily
available today) were beyond the manufacturing technology of
their times. Physicist David Miller recounts how a sophisticated
integrated circuit design project at TRW hit similar limits in
the early 1980s: "It all came down to whether a German
company could cool their glass lenses slowly enough to give us
the accuracy we needed. They couldn't."
In the molecular world, tool development again paces progress,
and new tools can bring breathtaking advances. Mark Pearson,
director of molecular biology for Du Pont, has observed this in
action: "When I was a graduate student back in the 1950s, it
was a multiyear problem to determine the molecular structure of a
single protein. We used to say, 'one protein, one career.' Yet
now the time has shrunk from a career to a decade to a
year—and in optimal cases to a few months." Protein
structures can be mapped atom by atom by studying X-ray
reflections from layers in protein crystals. Pearson observes
that "Characterizing a protein was a career-long endeavor in
part because it was so difficult to get crystals, and just
getting the material was a big constraint. With new technologies,
we can get our hands on the material now—that may sound
mundane, but it's a great advance. To the people in the field, it
makes all the difference in the world." Improved tools for
making and studying proteins are of special importance because
proteins are promising building blocks for first-generation
molecular machines.
But Isn't Science About Discoveries,
Not Tools?
Nobel Prizes are more often awarded for discoveries than for
the tools (including instruments and techniques) that made them
possible. If the goal is to spur scientific progress, this is a
shame. This pattern of reward extends throughout science, leading
to a chronic underinvestment in developing new tools. Philip
Abelson, an editor of the journal Science, points out that
the United States suffers from "a lack of support for
development of new instrumentation. At one time, we had a virtual
monopoly in pioneering advances in instrumentation. Now
practically no federal funds are available to universities for
the purpose." It's easier and less risky to squeeze one more
piece of data out of an existing tool than to pioneer the
development of a new one, and it takes less imagination.
But new tools emerge anyway, often from sources in other
fields. The study of protein crystals, for example, can benefit
from new X-ray sources developed by physicists, and techniques
from chemistry can help make new proteins. Because they can't
anticipate tools resulting from innovations in other fields,
scientists and engineers are often too pessimistic about what can
be achieved in their own fields. Nanotechnology will join several
fields, and yield tools useful in many others. We should expect
surprising results.
What Tools Do Researchers Use to Build
Small Devices?
Today's tools for making small-scale structures are of two
kinds: molecular-processing tools and bulk-processing tools. For
decades, chemists and molecular biologists have been using better
and better molecular-processing tools to make and manipulate
precise, molecular structures. These tools are of obvious use.
Physicists, as we will see, have recently developed tools that
can also manipulate molecules.
Combined with techniques from chemistry and molecular biology,
these physicist's tools promise great advances.
Microtechnologists have applied chip-making techniques to the
manufacture of microscopic machines. These technologies—the
main approach to miniaturization in recent decades—can play
at most a supporting role in the development of nanotechnology.
Despite appearances, it seems that microtechnology cannot be
refined into nanotechnology.
But Isn't Nanotechnology Just Very
Small Microtechnology?
For many years, it was conventional to assume that the road to
very small devices led through smaller and smaller devices: a
top-down path. On this path, progress is measured by
miniaturization: How small a transistor can we build? How small a
motor? How thin a line can we draw on the surface of a crystal?
Miniaturization focuses on scale and has paid off well, spawning
industries ranging from watchmaking to microelectronics.
Researchers at AT&T Bell Labs, the University of
California at Berkeley, and other laboratories in the United
States have used micromachining (based on microelectronic
technologies) to make tiny gears and even electric motors.
Micromachining is also being pursued successfully in Japan and
Germany. These microgears and micromotors are, however, enormous
by nanotechnological standards: a typical device is measured in
tens of micrometers, billions of times the volume of comparable
nanogears and nanomotors. (In our simulated molecular world, ten
microns is the size of a small town.) In size, confusing
microtechnology with molecular nanotechnology is like confusing
an elephant with a ladybug.
The differences run deeper, though. Microtechnology dumps
atoms on surfaces and digs them away again in bulk, with no
regard for which atom goes where. Its methods are inherently
crude. Molecular nanotechnology, in contrast, positions each atom
with care. As Bill DeGrado, a protein chemist at Du Pont, says,
"The essence of nanotechnology is that people have worked
for years making things smaller and smaller until we're
approaching molecular dimensions. At that point, one can't make
smaller things except by starting with molecules and building
them up into assemblies." The difference is basic: In
microtechnology, the challenge is to build smaller; in
nanotechnology, the challenge is to build bigger—we can
already make small molecules.
(A language warning: in recent years, nanotechnology has
indeed been used to mean "very small microtechnology";
for this usage, the answer to the above question is yes, by
definition. This use of a new word for a mere extension of an old
technology will produce considerable confusion, particularly in
light of the widespread use of nanotechnology in the sense
found here. Nanolithography, nanoelectronics,
nanocomposites, nanofabrication: not all that is nano- is molecular, or very
relevant to the concerns raised in this book. The terms molecular
nanotechnology and molecular
manufacturing are more awkward but avoid this
confusion.)
Will Microtechnology Lead to
Nanotechnology?
Can bulldozers can be used to make wristwatches? At most, they
can help to build factories in which watches are made. Though
there could be surprises, the relevance of microtechnology to
molecular nanotechnology seems similar. Instead, a bottom-up
approach is needed to accomplish engineering goals on the
molecular scale.
What Are the Main Tools Used for
Molecular Engineering?
Almost by definition, the path to molecular nanotechnology
must lead through molecular engineering. Working in different
disciplines, driven by different goals, researchers are making
progress in this field. Chemists are developing techniques able
to build precise molecular structures of sorts never before seen.
Biochemists are learning to build structures of familiar kinds,
such as proteins, to make new molecular objects.
In a visible sense, most of the tools used by chemists and
biochemists are rather unimpressive. They work on countertops
cluttered with dishes, bottles, tubes, and the like, mixing,
stirring, heating, and pouring liquids—in biochemistry, the
liquid is usually water with a trace of material dissolved in it.
Periodically, a bit of liquid is put into a larger machine and a
strip of paper comes out with a graph printed on it. As one might
guess from this description, research in the molecular sciences
is usually much less expensive than research in high-energy
physics (with its multibillion-dollar particle accelerators) or
research in space (with its multibillion-dollar spacecraft).
Chemistry has been called "small science," and not
because of the size of the molecules.
Chemists and biochemists advance their field chiefly by
developing new molecules that can serve as tools, helping to
build or study other molecules. Further advances come from new
instrumentation, new ways to examine molecules and determine
their structures and behaviors. Yet more advances come from new
software tools, new computer-based techniques for predicting how
a molecule with a particular structure will behave. Many of these
software tools let researchers peer through a screen into
simulated molecular worlds much like those toured in the last two
chapters.
Of these fields, it is biomolecular science that is most
obviously developing tools that can build nanotechnology, because
biomolecules already form molecular machines, including devices
resembling crude assemblers.
This path is easiest to picture, and can surely work, yet there
is no guarantee that it will be fastest: research groups
following another path may well win. Each of these paths is being
pursued worldwide, and on each, progress is accelerating.
Physicists have recently contributed new tools of great
promise for molecular engineering. These are the proximal probes,
including the scanning
tunneling microscope (STM) and the atomic
force microscope (AFM). A proximal-probe device places a
sharp tip in proximity to a surface and uses it to probe (and
sometimes modify) the surface and any molecules that may be stuck
to it.
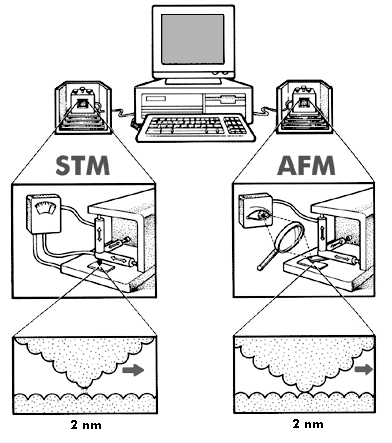
Figure 4: STM/AFM
The scanning tunneling microscope (STM, on the left) images
surfaces well enough to show individual atoms, sensing
surface contours by monitoring the current jumping the gap
between tip and surface. The atomic force microscope (AFM, on
the right) senses surface contours by mechanical contact,
drawing a tip over the surface and optically sensing its
motion as it passes over single-atom bumps.
How Does an STM Work?
An STM brings a sharp, electrically conducting needle up to an
electrically-conducting surface, almost touching it. The
needle and surface are electrically connected (see the left-hand
side of Figure 4), so that a current will flow if they touch,
like closing a switch. But at just what point do soft, fuzzy
atoms "touch"? It turns out that a detectable current
flows when just two atoms are in tenuous contact—fuzzy
fringes barely overlapping—one on the surface and one on the
tip of the needle. By delicately maneuvering the needle around
over the surface, keeping the current flowing at a tiny, constant
rate, the STM can map shape of the surface with great precision.
Indeed, to keep the current constant, the needle has to go up and
down as it passes over individual atoms.
The STM was invented by Gerd
Binnig and Heinrich
Rohrer, research physicists studying surface phenomena at
IBM's research labs in Zurich, Switzerland. After working through
the 1970s, Rohrer and Binnig submitted their first patent
disclosure on an STM in mid-1979. In 1982, they produced images
of a silicon surface, showing individual atoms. Ironically, the
importance of their work was not immediately recognized: Rohrer
and Binnig's first scientific paper on the new tool was rejected
for publication on the grounds that it was "not interesting
enough." Today, STM conferences draw interested researchers
by the hundreds from around the world.
In 1986—quite promptly as these things go—Binnig and
Rohrer were awarded a Nobel Prize. The Swedish Academy
explained its reasoning: "The scanning tunneling microscope
is completely new and we have so far seen only the beginning of
its development. It is, however, clear that entirely new fields
are opening up for the study of matter." STMs are no longer
exotic: Digital Instruments of Santa Barbara, California, sells
its system (the NanoscopeŽ) by mail with an
atomic-resolution-or-your-money-back guarantee. Within three
years of their commercial introduction, hundreds of STMs had been
purchased.
How Does an AFM Work?
The related atomic force microscope (on the right side of
Figure 4) is even simpler in concept: A sharp probe is dragged
over the surface, pressed down gently by a straight spring. The
instrument senses motions in the spring (usually optically), and
the spring moves up and down whenever the tip is dragged over an
atom on the surface. The tip "feels" the surface just
like a fingertip in the simulated molecular world. The AFM was
invented by Binnig, Quate, and Gerber at Stanford University and
IBM San Jose in 1985. After the success of the STM, the
importance of the AFM was immediately recognized. Among other
advantages, it works with nonconducting materials. The next
chapter will describe how AFM-based devices might be used as molecular manipulators
in developing molecular nanotechnology. As this is written, AFMs
have just become commercially available.
(Note that that AFMs and STMs are not quite as easy to use as
these descriptions might suggest. For example, a bad tip or a bad
surface can prevent atomic resolution, and pounding on the table
is not recommended when such sensitive instruments are in
operation. Further, scientists often have trouble deciding just
what they're seeing, even when they get a good image.)
Can Proximal Probes Move Atoms?
To those thinking in terms of nanotechnology, STMs immediately
looked promising not only for seeing atoms and molecules but for
manipulating them. This idea soon became widespread among
physicists. As Calvin Quate stated in Physics Today in
1986, "Some of us believe that the scanning tunneling
microscope will evolve . . . that one day [it] will be used to
write and read patterns of molecular size." This approach
was suggested as a
path to molecular nanotechnology in Engines of Creation,
again in 1986.
By now, whole stacks of scientific papers document the use of
STM and AFM tips to scratch, melt, erode, indent, and otherwise
modify surfaces on a nanometer scale. These operations move atoms
around, but with little control. They amount to bulk operations
on a tiny scale— one fine scratch a few dozen atoms
wide, instead of the billions that result from conventional
polishing operations.
Can Proximal Probes Move Atoms More
Precisely?
In 1987, R. S. Becker, J. A. Golovchenko, and B. S.
Swartzentruber at AT&T Bell Laboratories announced that they
had used an STM to deposit small blobs on a germanium surface.
Each blob was thought to consist of one or a few germanium atoms.
Shortly thereafter, IBM Almaden researchers John Foster, Jane
Frommer, and Patrick Arnett achieved a milestone in STM-based
molecular manipulation. Of this team, Foster and Arnett attended
the First Foresight
Conference on Nanotechnology, where they told us the
motivations behind their work.
Foster came to IBM from Stanford University, where he had
completed a doctorate in physics and taught at graduate school.
The STM work was one of his first projects in the corporate
world. He describes his colleague Arnett as a former
"semiconductor jock" involved in chip creation at IBM's
Burlington and Yorktown locations. Besides his doctorate in
physics, Arnett brought mechanical-engineering training to the
effort.
Arnett explains what they were trying to do: "We wanted
to see if you could do something on an atomic scale, to create a
mechanism for storing information and getting it back
reliably." The answer was yes. In January 1988, the journal Nature
carried their letter reporting success in pinning an organic
molecule to a particular location on a surface, using an STM to
form a chemical bond by applying an electrical pulse through the
tip. They found that having created and sensed the feature, they
could go back and use another voltage pulse from the tip to
change the feature again: enlarging it, partly erasing it, or
completely removing it.
IBM quickly saw a commercial use, as explained by Paul M.
Horn, acting director of physical sciences at the Thomas J.
Watson Research Center: "This means you can create a storage
element the size of an atom. Ultimately, the ability to do that
could lead to storage that is ten million times more dense than
anything we have today." A broader vision was given by
another researcher, J. B. Pethica, in the issue of Nature
in which the work appeared: "The partial erasure reported by
Foster et al. implies that molecules may have pieces
deliberately removed, and in principle be atomically 'edited,'
thereby demonstrating one of the ideals of nanotechnology."
Can Proximal Probes Move Atoms With
Complete Precision?
Foster's group succeeded in pinning single molecules to a
surface, but they couldn't control the results—the position
and orientation—precisely. In April 1990, however, another
group at the same laboratory carried the
manipulation of atoms even further, bringing a splash of
publicity. Admittedly, the story must have been hard to resist:
it was accompanied by an STM picture of the name IBM,"
spelled out with thirty-five precisely placed atoms (Figure 5).
The precision here is complete, like the precision of molecular
assembly: each atom sits in a dimple on the surface of a nickel
crystal; it can rest either in one dimple or in another, but
never somewhere between.

FIGURE 5: WORLD SMALLEST LOGO—35 XENON ATOMS (Courtesy of
IBM Research Division)
Donald
Eigler, the lead author on the Nature paper describing
this work, sees clearly where all this is leading: "For
decades, the electronics industry has been facing the challenge
of how to build smaller and smaller structures. For those of us
who will now be using individual atoms as building blocks, the
challenge will be how to build up structures atom by atom."
How Far Can Proximal Probes Take Us?
Proximal probes have advantages as a tool for developing
nanotechnology, but also weaknesses. Today, their working tips
are rough and irregular, typically even rougher than shown in
Figure 4. To make stable bonds form, John Foster's group used a
pulse of electricity, but the results proved hard to control. The
"IBM" spelled out by Donald Eigler's group was precise,
but stable only at temperatures near absolute zero—such
patterns vanish at room temperature because they are not based on
stable chemical bonds. Building structures that are both stable
and precise is still a challenge. To form stable bonds in precise
patterns is the next big challenge.
John Foster says, "We're exploring a concept which we
call 'molecular herding,' using the STM to 'herd' molecules the
way my Shetland sheep dog would herd sheep . . . Our ultimate
goal with molecular herding is to make one particular molecule
move to another particular one, and then essentially force them
together. If you could put two molecules that might be small
parts of a nanomachine on
the surface, then this kind of herding would allow you to haul
one of them up to the other. Instead of requiring random motion
of a liquid and specific chemical lock-and-key interactions to
give you exactly what you want in bringing two molecules together
[as in chemical and biochemical approaches], you could drive that
reaction on a local level with the STM. You could use the STM to
put things where you want them to be." The next chapter will
discuss additional ideas for using proximal probes in early
nanotechnology.
Proximal-probe instruments may be a big help in building the
first generation of nanomachines, but they have a basic limit:
Each instrument is huge on a molecular scale, and each could bond
only one molecular piece at a time. To make anything
large—say, large enough to see with the naked eye—would
take an absurdly long time. A device of this sort could add one
piece per second, but even a pinhead contains more atoms than the
number of seconds since the formation of Earth. Building a Pocket
Library this way would be a long-term project.
How Can Such Slow Systems Ever Build
Anything Big?
Rabbits and dandelions contain structures put together one
molecular piece at a time, yet they grow and reproduce quickly.
How? They build in parallel, with many billions of molecular
machines working at once. To gain the benefits of such enormous
parallelism, researchers can either 1) use proximal probes to
build a better, next-generation technology, or 2) use a different
approach from the start.
The techniques of chemistry and biomolecular engineering
already have enormous parallelism, and already build precise
molecular structures. Their methods, however, are less direct
than the still hypothetical proximal probe-based
molecule-positioners. They use molecular building blocks shaped
to fit together spontaneously, in a process of self-assembly.
David Biegelsen, a physicist who works with STMs at the Xerox
Palo Alto Research Center, put it this way at the nanotechnology
conference: "Clearly, assembly using STMs and other variants
will have to be tried. But biological systems are an existence
proof that assembly and self-assembly can be done. I don't see
why one should try to deviate from something that already exists.
What Are the Main Advantages of
Molecular Building Blocks?
A huge technology base for molecular construction already
exists. Tools originally developed by biochemists and
biotechnologists to deal with molecular machines found in nature
can be redirected to make new molecular machines. The expertise
built up by chemists in more than a century of steady progress
will be crucial in molecular design and construction. Both
disciplines routinely handle molecules by the billions and get
them to form patterns by self-assembly. Biochemists, in
particular, can begin by copying designs from nature.
Molecular building-block strategies could work together with
proximal probe strategies, or could replace them, jumping
directly to the construction of large numbers of molecular
machines. Either way, protein molecules are likely to play a
central role, as they do in nature.
How Can Protein Engineering Build
Molecular Machines?
Proteins can self assemble into working molecular machines,
objects that do something, such as cutting and splicing
other molecules or making muscles contract. They also join with
other molecules to form huge assemblies like the ribosome (about the size of a
washing machine, in our simulation view).
Ribosomes—programmable machines for manufacturing
proteins—are nature's closest approach to a molecular
assembler. The genetic-engineering industry is chiefly in the
business of reprogramming natural nanomachines, the ribosomes, to
make new proteins or to make familiar proteins more cheaply.
Designing new proteins is termed protein
engineering. Since biomolecules already form such complex
devices, it's easy to see that advanced protein engineering could
be used to build first-generation nanomachines.
If We Can Make Proteins, Why Aren't We
Building Fancy Molecular Machines?
Making proteins is easier than designing them. Protein
chemists began by studying proteins found in nature, but have
only recently moved on to the problem of engineering new ones.
These are called de novo proteins, meaning completely new,
made from scratch. Designing proteins is difficult because of the
way they are constructed. As Bill
DeGrado, a protein chemist at Du Pont, explains: "A
characteristic of proteins is that their activities depend on
their three-dimensional structures. These activities may range
from hormonal action to a function in digestion or in metabolism.
Whatever their function, it's always essential to have a definite
three-dimensional shape or structure." This
three-dimensional structure forms when a chain folds to form a
compact molecular object. To get a feel for how tough it is to
predict the natural folding of a protein chain, picture a
straight piece of cord with hundreds of magnets and sticky knots
along its length. In this state, it's easy to make and easy to
understand. Now pick it up, put it in a glass jar, and shake it
for a long time. Could you predict its final shape? Certainly
not: it's a tangled mess. One might call this effort at
prediction "the sticky-cord-folding problem"; protein
chemists call theirs "the protein-folding problem."
Given the correct conditions, a protein chain always folds
into one special shape, but that shape is hard to predict from
just the straightened structure. Protein designers, though, face
the different job of first determining a desired final shape, and
then figuring out what linear sequence of amino acids to use to
make that shape. Without solving the classic protein-folding
problem, they have begun to solve the protein-design
problem.
What Has Been Accomplished So Far?
Bill DeGrado and his colleagues at Du Pont had one of the
first successes: "We've been able to use basic principles to
design and build a simple molecule that folds up the way we want
it to. This is really the first real example of a designed
protein structure, designed from scratch, not by taking an
already existing structure and tinkering with it."
Although scientists do the work, the work itself is really a
form of engineering, as shown by the title of the field's
journal, Protein Engineering. Bill DeGrado's description
of the process makes this clear: "After you've made it, the
next step is to find out whether your protein did what you
expected it to do. Did it fold? Did it pass ions across bilayers
[such as cell membranes]? Does
it have a catalytic function [speeding specific chemical
reactions]? And that's tested using the appropriate experiment.
More than likely, it won't have done what you wanted it to do, so
you have to find out why. Now, a good design has in it a
contingency plan for failure and helps you learn from mistakes.
Rather than designing a structure that would take a year or more
to analyze, you design it so that it can be assayed for given
function or structure in a matter of days."
Many groups are pursuing protein
design today, including academic researchers like Jane and
Dave Richardson at Duke University, Bruce Erickson at the
University of North Carolina, and Tom Blundell, Robin
Leatherbarrow, and Alan Fersht in Britain. The successes have
started to roll in. Japan, however, is unique in having an
organization devoted exclusively to such projects: the Protein
Engineering Research Institute (PERI) in Osaka. In 1990, PERI
announced the successful design and construction of a de novo protein
several times larger than any built before.
Is There Anything Special About
Proteins?
The main advantage of proteins is that they are familiar: a
lot is known about them, and many tools exist for working with
them. Yet proteins have disadvantages as well. Just because this
design work is starting with proteins—soft, squishy
molecules that are only marginally suitable for
nanotechnology—doesn't mean it will stay within those
limits. De Grado points out "The fundamental goal of our
work in de novo design is to be able to take the next step
and get entirely away from protein systems." An early
example is the work of Wallace Carothers of Du Pont, who used a de
novo approach to studying the nature of proteins: Rather than
trying to cut up proteins, he tried to build up things starting
with amino acids and other similar monomers. In 1935, he
succeeded in making nylon.
DeGrado explains "There is a deep philosophical belief at
Du Pont in the ability of people to make molecules de novo
that will do useful things. And there is a fair degree of
commitment from the management that following that path will lead
to products: not directly, and not always predictably, but they
know that they need to support the basic science.
"I think ultimately we have a better chance at doing some
really exciting things by de novo design, because our
repertory should be much greater than that of nature. Think about
the ability to fly: One could breed better carrier pigeons or one
could design airplanes." The biology community, however,
leans more toward ornithology than toward aerospace engineering.
DeGrado's experience is that "a lot of biologists feel that
if you aren't working with the real thing [natural proteins], you
aren't studying biology, so they don't totally accept what we're
doing. On the other hand, they recognize it as good
chemistry."
Where Is Protein Engineering Headed?
Like the IBM physicists, protein designers are moved by a
vision of molecular engineering. In 1989, Bill DeGrado predicted,
"I think we'll be able to make catalysts or enzymelike
molecules, possibly ones that catalyze reactions not catalyzed in
nature." Catalysts are molecular machines that speed up
chemical reactions: they form a shape for the two reacting
molecules to fit into and thereby help the reaction move faster,
up to a million reactions per second. New ones, for reactions
that now go slowly, will give enormous cost savings to the
chemical industry.
This prediction was borne out just a few months later, when
Denver researchers John Stewart, Karl Hahn, and Wieslaw Klis
announced their new enzyme, designed from scratch over a period
of two years and built successfully on the first try. It's a
catalyst, making some reactions go about 100,000 times faster.
Nobel Prize-winning biochemist Bruce Merrifield believes that
"if others can reproduce and expand on this work, it will be
one of the most important achievements in biology or
chemistry."
DeGrado also has longer term plans for protein design, beyond
making new catalysts: "It will allow us to think about
designing molecular devices in the next five to ten years. It
should be possible ultimately to specify a particular design and
build it. Then you'll have, say, proteinlike molecules that
self-assemble into complex molecular objects, which can serve as
machinery. But there's a limit to how small you can make devices.
You'll shrink things down so far and then you won't be able to go
any further, because you've reached molecular dimensions."
Mark Pearson shows that management at Du Pont also has this
vision. Regarding the prospects for nanotechnology and
assemblers, he remarked, "You know, it'll take money and
effort and good ideas for sure. But to my way of thinking, there
is no absolute fundamental limitation to preclude us from doing
this kind of thing." He didn't say his company plans to
develop nanotechnology, but such plans aren't really necessary.
Du Pont is already on the nanotechnology path, for
other—shorter-term, commercial—reasons. Like IBM, if
they do decide to move quickly, they have the resources and
forward-looking people needed to succeed.
Who Else Builds Molecular Objects?
Chemists, most of whom do not work on proteins, are the
traditional experts in building molecular objects. As a group
they've been building molecules for over a century, with ever
increasing ability and confidence. Their methods are all
indirect: They work with billions of atoms at a time—massive
parallelism—but without control of the positions of their
workpieces. The molecules typically tumble randomly in a liquid
or gas, like pieces of a puzzle that may or may not fit together
correctly when shaken together in a box. With clever design and
planning, most pieces will join properly.
Chemists mix molecules on a huge scale (in our simulation
view, a test tube holds a churning molecular swarm with the
volume of an inland sea), yet they still achieve precise
molecular transformations. Given that they work so indirectly,
their achievements are astounding. This is, in part, the result
of the enormous amount of work poured into the field for many
decades. Thousands of chemists are working on molecular
construction in the United States alone; add to that the chemists
in Europe, in Japan, and in the rest of the world, and you have a
huge community of researchers making great strides. Though it
publishes only a one-paragraph summary of each research report, a
guide to the chemical literature—Chemical Abstracts—covers
several library walls and grows by many feet of shelf space every
year.
How Can Mixing Chemicals Build
Molecular Objects?
An engineer would say that chemists (at least those
specializing in synthesis) are doing construction work, and would
be amazed that they can accomplish anything without being able to
grab parts and put them in place. Chemists, in effect, work with
their hands tied behind their backs. Molecular manufacturing can
be termed "positional chemistry" or "positional synthesis,"
and will give chemists the ability to put molecules where they
want them in three-dimensional space. Rather than trying to
design puzzle pieces that will stick together properly by
themselves when shaken together in a box, chemists will then be
able to treat molecules more like bricks to be stacked. The basic
principles of chemistry will be the same, but strategies for
construction will become far simpler.
Without positional control, chemists face a problem something
like this: Picture a giant glass barrel full of tiny
battery-powered drills, buzzing away in all directions, vibrating
around in the barrel. Your goal is to take a piece of wood and
put a hole in just one specific spot. If you simply throw it in
the barrel, it will be drilled haphazardly in many places. To
control the process, you must protect all the places you don't
want drilled—perhaps by gluing protective pieces of metal
over most of the wood surface. This problem—how to protect
one part of a molecule while altering another part—has
forced chemists to develop ever-cleverer ploys to build larger
and larger molecules.
If Chemists Can Make Molecules, Why
Aren't They Building Fancy Molecular Machines?
Chemists can achieve great things, but have focused much of
their effort on duplicating molecules found in nature and then
making minor variants. As an example, take palytoxin, a molecule
found in a Hawaiian coral. It was so difficult to make in the lab
that it has been called "the Mount Everest of synthetic
chemistry," and its synthesis was hailed as a triumph. Other
efforts are poured into making small molecules with unusual
bonding, or molecules of remarkable symmetry, like
"cubane" and "dodecahedrane" (shaped like the
Platonic solids they are named after).
Chemists, at least in the United States, regard themselves as
natural scientists even when their life's work is the
construction of molecules by artificial means. Ordinarily, people
who build things are called engineers. And indeed, at the
University of Tokyo the Department of Synthetic Chemistry is part
of the Faculty of Engineering; its chemists are designing
molecular switches for storing computer data. Engineering
achievements will require work directed at engineering goals.
How Could Chemists Move Toward Building
Molecular Machines?
Molecular engineers working toward nanotechnology need a set
of molecular building blocks for making large, complex
structures. Systematic building-block construction was pioneered
by Bruce
Merrifield, winner of the 1984
Nobel Prize in Chemistry. His approach, known as "solid
phase synthesis," or simply "the Merrifield
method," is used to synthesize the long chains of amino
acids that form proteins. In the Merrifield method, cycles of
chemical reaction each add one molecular building block to the
end of a chain anchored to a solid support. This happens in
parallel to each of trillions of identical chains, building up
trillions of molecular objects with a particular sequence of
building blocks. Chemists routinely use the Merrifield method to
make molecules larger than palytoxin, and related techniques are
used for making DNA in so-called
gene machines: an ad from an Alabama company reads, "Custom
DNA—Purified and Delivered in 48 hours."
While it's hard to predict how a natural protein chain will
fold—they weren't designed to fold predictably—chemists
could make building blocks that are larger, more diverse, and
more inclined to fold up in a single, obvious, stable pattern.
With a set of building blocks like these, and the Merrifield
method to string them together, molecular engineers could design
and build molecular machines with greater ease.
How Do Researchers Design What They
Can't See?
To make a new molecule, both its structure and the procedure
to make it must be designed. Compared to gigantic science
projects like the Superconducting Supercollider and the Hubble
Space Telescope, working with molecules can be done on a
shoestring budget. Still, the costs of trying many different
procedures add up. To help predict in advance what will work and
what won't, designers turn to models.
You may have played with molecular models in chemistry class:
colored plastic balls and sticks that fit together like Tinker
Toys. Each color represents a different kind of atom: carbon,
hydrogen, and so on. Even simple plastic models can give you a
feel for how many bonds each kind of atom makes, how long the
bonds are, and at what angles they are made. A more sophisticated
form of model uses only spheres and partial spheres, without
sticks. These colorful, bumpy shapes are called CPK models, and
are widely used by professional chemists. Nobel laureate Donald
Cram remarks that "We have spent hundreds of hours
building CPK models of potential complexes and grading them for
desirability as research targets." His research, like that
of fellow Nobelists Charles
J. Pedersen and Jean-Marie
Lehn, has focused on designing and making medium-sized
molecules that self assemble.
Although physical models can't give a good description of how
molecules bend and move, computer-based molecules can.
Computer-based modeling is already playing a key role in
molecular engineering. As John Walker (a founder and leader of
Autodesk) has remarked, "Unlike all of the industrial
revolutions that preceded it, molecular engineering requires, as
an essential component, the ability to design, model, and
simulate molecular structures using computers."
This has not gone unnoticed in the business community. John
Walker's remark was part of a talk on nanotechnology given at
Autodesk, a leader in computer-aided design and one of the five
largest software firms in the United States. Soon after this
talk, the company made its first major investment in the
computer-aided design of molecules.
[See Nanotechnology
in Manufacturing, by John Walker]
How Does Molecular Design Compare to
More Familiar Kinds of Engineering?
Manufacturers and architects know that designs for new
products and buildings are best done on a computer, by
computer-aided design (CAD). The new molecular design software
can be called molecular CAD, and in its forefront are
researchers such as Jay Ponder
of the Yale University Department of Molecular Biophysics and
Biochemistry. Ponder explains that "There's a strong link
between what molecular designers are doing and what architects
do. Michael
Ward of Du Pont is designing a set of building blocks for a
Tinker Toy set so that you can build larger structures. That's
exactly what we're doing with molecular modeling techniques.
"All the design and mechanical engineering principles
that apply to building a skyscraper or a bridge apply to
molecular architecture as well. If you're building a bridge,
you're going to model it and see how many trucks can be on the
bridge at the same time without it collapsing, what kind of
forces you're going to apply to it, whether it can stand up to an
earthquake.
"And the same process goes on in molecular design: You're
designing pieces and then analyzing the stresses and forces and
how they will change and perturb the structure. It's exactly the
same as designing and building a building, or analyzing the
stresses on any macroscale structure. I think it's important to
get people to think in those terms.
"The molecular designer has to be creative in the same
way that an architect has to be creative in designing a building.
When people are looking at the interior of a protein structure
and trying to redesign it to create a space that will have a
particular function, such as binding to particular molecules,
that's like designing a room to use as a dining room—one
that will fit certain sizes of tables and certain numbers of
guests. It's the same thing in both cases: You have to design a
space for a function."
Ponder combines chemistry and computer science with an overall
engineering approach: "I'm kind of a hybrid. I spend about
half my time doing experiments and about half my time writing
computer programs and doing computational work. In the
laboratory, I create or design molecules to test some of the
computational ideas. So I'm at the interface." The
engineering perspective helps in thinking about where molecular
research can lead: "Even though with nanotechnology we're at
the nanometer scale, the structures are still big enough that an
awful lot of things are classical. Again, it's really like
building bridges—very small bridges. And so there are many
almost standard mechanical-engineering techniques for
architecture and building structures, such as stress analysis,
that apply."
Doesn't Engineering Require More
Teamwork Than Science Does?
Getting to nanotechnology will require the work of experts in
differing fields: chemists, who are learning how to make
molecular machines; computer scientists, who are building the
needed design tools; and perhaps STM and AFM experts, who can
provide tools for molecular positioning. To make progress,
however, these experts must do more than just work, they must
work together. Because nanotechnology is inherently
interdisciplinary, countries that draw hard lines between their
academic disciplines, as the United States does, will find that
their researchers have difficulty communicating and cooperating.
In chemistry today, a half-dozen researchers aided by a few
tens of students and technicians is considered a large team. In
aerospace engineering, enormous tasks like reaching the Moon or
building a new airliner are broken down into tasks that are
within the reach of small teams. All these small teams work
together, forming a large team that may consist of thousands of
engineers aided by many thousands of technicians. If chemistry is
to move in the direction of molecular
systems engineering, chemists will need to take at least a
few steps in this direction.
In engineering, everyone knows that designing a rocket will
require skills from many disciplines. Some engineers know
structures, others know pumps, combustion, electronics, software,
aerodynamics, control theory, and so on and so forth down a long
list of disciplines. Engineering managers know how to bring
different disciplines together to build systems.
In academic science, interdisciplinary work is productive and
praised, but is relatively rare. Scientists don't need to
cooperate to have their results fit together: they are all
describing different parts of the same thing—nature—so
in the long run, their results tend to come together into a
single picture. Engineering, however, is different. Because it is
more creative (it actually creates complex things), it
demands more attention to teamwork. If the finished parts are
going to work together, they must be developed by groups that
share a common picture of what each part must accomplish.
Engineers in different disciplines are forced to communicate; the
challenge of management and team-building is to make that
communication happen. This will apply to engineering molecular
systems as much as it does to engineering computers, cars,
aircraft, or factories.
Jay Ponder suggests that it's a question of perspective.
"It's all a matter of what's perceived to be important by
the different groups that have to come together to make this
work: the chemists doing their bit and the computational people
doing their bit. People have to come together and see the big
picture. There are people who try to bridge the gaps, but they
are rare compared to the people who just work in their own
specialty." Progress toward nanotechnology will continue,
and as it does, researchers trained as chemists, physicists, and
the like will learn to talk to one another to solve new problems.
They will either learn to think like engineers and work in teams,
or they will be eclipsed by colleagues who do.
Are These Problems Preventing Advances?
With all these problems, the advance toward nanotechnology
steadily continues. Industry must gain ever-better control of
matter to stay competitive in the world marketplace. The STM,
protein engineering, and much of chemistry are driven by
commercial imperatives. Focused efforts would yield faster
advances, yet even without a clear focus, advances in this
direction have an air of inevitability. As Bill DeGrado observes,
"We really do have the tools. Experience has shown that when
you have the analytic and synthetic tools to do things, in the
end science goes ahead and does them—because they are
doable." Jay Ponder agrees: "Over the next few years,
you're going to see slow evolutionary advances coming from people
tinkering with molecular structures and figuring out their
principles. People are going to work on a particular problem
because they see some application for it or because they got
grant funding for it. And in the process of doing something like
improving a laundry detergent's ability to clean protein stains,
Proctor and Gamble is going to help work out the principles for
how to increase molecular stability, and to design spaces inside
the molecules."
Are the Japanese Bearing Their Share of
the Burden in Nanotechnology Research?
For a variety of reasons, Japan's contribution to
nanotechnology research promises to be excellent. While the
United States has generally pursued researching this area with
little sense of long-term direction, it appears that Japan has
begun to take a more focused approach. Researchers there already
have clear ideas about molecular machines—about what might
work and what probably won't. Japanese researchers are accustomed
to a higher level of interdisciplinary contact and engineering
emphasis than are Americans. In the United States, we prize
"basic science," often calling it "pure
science," as if to imply that practical applications are a
form of impurity. Japan instead emphasizes "basic
technology."
Nanotechnology is a basic technology, and the Japanese
recognize it as such. Recent changes at the Tokyo Institute of
Technology—Japan's equivalent of MIT—reflect their
views of promising directions for future research. For many
decades, Tokyo Tech has had two major divisions: a Faculty of
Science and a Faculty of Engineering. To these is now being added
a Faculty of Bioscience and Biotechnology, to consist of four
departments: a Department of Bioscience, a Department of
Bioengineering, a Department of Biomolecular Engineering, and
what is termed a "Department of Biostructure." The
creation of a new faculty in a major Japanese university is a
rare event. What U.S. university has a department explicitly
devoted to molecular engineering? Japan has both the departments
at Tokyo Tech and Kyoto University's recently established
Department of Molecular Engineering.
Japan's Institute for Physical and Chemical Research (RIKEN)
has broad-based interdisciplinary strength. Hiroyuki Sasabe, head
of the Frontier Materials Research Program at RIKEN, notes that
the institute has expertise in organic synthesis, protein
engineering, and STM technology. Sasabe says that his laboratory
may need a molecular manipulator of the sort described in the
next chapter to accomplish its goals in molecular engineering.
Research consortia in Japan are also moving toward
nanotechnology. The Exploratory
Research for Advanced Technology Organization (ERATO)
sponsors many three-to-five year projects in parallel, each with
a specific goal. Consider the work in progress:
• Yoshida
Nanomechanism Project
• Hotani
Molecular Dynamic Assembly Project
• Kunitake
Molecular Architecture Project
• Nagayama
Protein Array Project
• Aono
Atomcraft Project
These focus on different aspects of gaining control over
matter at the atomic level. The Nagayama Protein Array Project
aims to use proteins as engineering materials to move toward
making new molecular devices. The Aono Atomcraft Project does not
involve nuclear power—as its translation might
imply—but is instead an interdisciplinary effort to use an
STM to arrange matter on the atomic scale.
At some point, work on nanotechnology must move beyond
spin-offs from other fields and undertake the design and
construction of molecular machinery. This shift from
opportunistic science to organized engineering requires a change
in attitude. In this, Japan leads the United States.
What Is a Good Educated Guess of How
Long It Will Take to Develop Molecular Nanotechnology?
Molecular nanotechnology will emerge step by step. Major
milestones, such as the engineering of proteins and the
positioning of individual atoms, have already been passed. To get
a sense of the likely pace of developments, we need to look at
how various trends fit together.
Computer-based molecular-modeling tools are spawning
computer-aided design tools. These will grow more capable. The
underlying technology base—computer hardware—has for
decades been improving in price and performance on a steeply
rising curve, which is generally expected to continue for many
years. These advances are quite independent of progress in
molecular engineering, but they make molecular engineering
easier, speeding advances. Computer models of
molecular machines are beginning to appear, and these will
whet the appetites of researchers.
Progress in engineering molecular machines, whether using
proximal probes or self-assembly, will eventually achieve
striking successes; the objectives of research in Japan will
begin to draw serious attention; understanding of the long-term
promise of molecular engineering will become more widespread.
Some combination of these developments will eventually lead to a
serious, public appraisal of what these technologies can
achieve—and then the world of opinion, funding, and research
fashion will change. Before, advances will be steady but
haphazard; afterward, advances will be driven with the energy
that flows into major commercial, military, and medical research
programs, because nanotechnology will be recognized as furthering
major commercial, military, and medical goals. The timing of
subsequent events depends largely on when this threshold of
serious attention is reached.
In making time estimates, people are prone to assume that a
large change must take a long time. Most do, but not all. Pocket
calculators had a dramatic effect on the slide-rule industry:
they replaced it. The speed of this change caught the slide rule
moguls by surprise, but the pace of progress in electronics
didn't slow down merely to suit their expectations.
One can argue that nanotechnology will be developed fast: many
countries and companies will be competing to get there first.
They will be driven onward both by the immense expected
benefits—in many areas, including medicine and the
environment—as well as by potential military applications.
That is a powerful combination of motives, and competition is a
powerful accelerator.
A counterargument, though, suggests that development will be
slow: anyone who has done anything of significance in the real
world of technology—doing a scientific experiment, writing a
computer program, bringing a new product to market—knows
that these goals take longer than expected. Indeed, Hofstadter's
Law states that projects take longer than expected, even when
Hofstadter's Law is taken into account. This principle is a good
guide for the short term, and for a single project.
The situation differs, though, when many different approaches
are being explored by many different groups over a period of
years. Most projects may take longer than expected, but with many
teams trying many approaches, one approach may prove faster than
expected. The winner of a race is always faster than the average
runner. John Walker notes, "The remarkable thing about
molecular engineering is that it looks like there are many
different ways to get there and, at the moment, rapid progress is
being made along every path—all at the same time."
Also, technology development is like a race run over an
unmapped course. When the first runners reach the top of a hill,
they may see a shortcut. A trailing runner may decide to crash
off into the bushes, and stumble across a bicycle and a paved
road. The progress of technology is seldom predictable because
progress often reveals new directions.
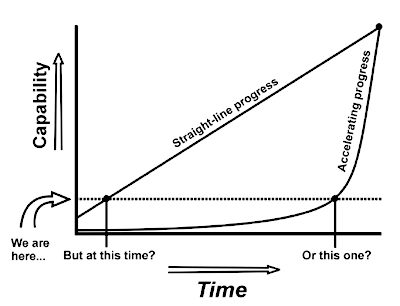
GRAPH OF LINEAR VS. ACCELERATING GROWTH OF TECHNOLOGY
How close we are to goal depends on whether technological
advances are a constant pace of accelerating. In this diagram,
the dashed line represents the current level of technology, and
the large dot in the upper right represents a goal such as
nanotechnology. With a straight-line advance, it's easier to
estimate how far away a goal is. With an accelerating advance, a
goal can be reached with little warning.
So how can we estimate a date for the arrival of
nanotechnology? It's safest to take a cautious approach: When
anticipating benefits, assume it's far off; when preparing for
potential problems, assume it's right around the corner. The old
folk saying applies: Hope for the best, prepare for the worst.
Any dates assigned to "far off" and "right around
the corner" can be no better than educated
guesses—molecular behavior can be calculated, but not
technology timetables of this sort. With those caveats, we would
estimate that general-purpose molecular assemblers will likely be
developed in the early decades of the twenty-first century,
perhaps in the first.
John Walker, whose technological foresight has led Autodesk
from start-up to a dominant role in its industry, points out that
not long ago, "Many visionaries intimately familiar with the
developments of silicon technology still forecast it would take
between twenty and fifty years before molecular engineering
became a reality. This is well beyond the planning horizon of
most companies. But recently, everything has begun to
change." Based on the new developments, Walker places his
bet: "Current progress suggests the revolution may happen
within this decade, perhaps starting within five years.
|